ALBUQUERQUE, N.M. — The genome microarray studies being done by Sandia National Laboratories and the University of New Mexico (UNM) rely on two major enabling technologies: the arrays themselves and the scanners that read the information from the arrays.
The Keck-funded research uses commercially available arrays from Affymetrix, Inc., but laboratory arrayers give researchers the option to build custom arrays and to work with organisms for which commercial arrays are not yet available.
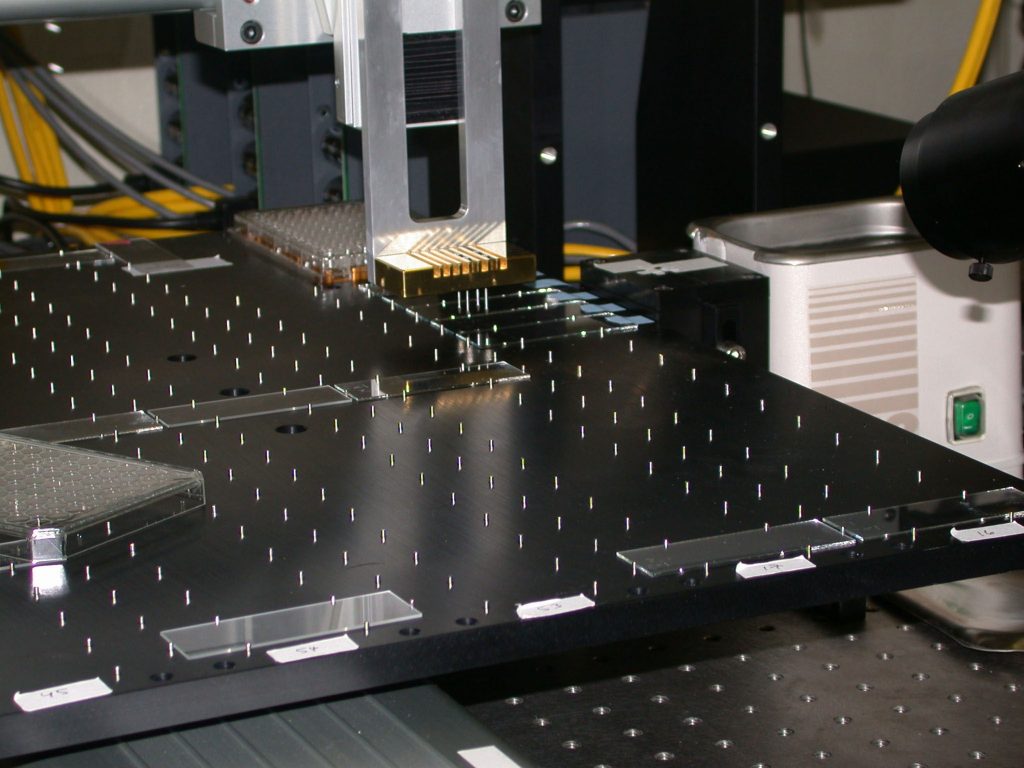
“One of our first steps was to get access to an arrayer. We ordered the parts and built it over about a week with a group of undergraduates,” says Sandian George Davidson. “This was a nice example of the benefits of collaborating with a ‘can-do’ organization like Sandia.”
This initial arrayer has just been replaced with a faster commercial machine, but it served as the main research tool for developing all of the arraying protocols that are now standard in biology professor Maggie Werner-Washburne’s laboratory at UNM.
The instrument is a robot, which moves metal pins, first dipping them into reservoirs of DNA, one gene per reservoir, and then tapping the nanoliter quantities onto the surfaces of up to 100 glass slides. Thousands of spots containing different DNA genes are laid out in each array on 100-200 micrometer centers.
After a slide is printed, samples of RNA from the target cells are prepared and translated into DNA complementary to the gene that originally coded the RNA molecule. These cDNA molecules are tagged with a fluorescent marker and incubated above the spots of DNA on the array.
The cDNA find the corresponding gene printed on the array and bind together in a process called competitive hybridization. Interrogation of the array with a fluorescent scanning device measures the concentration of the cDNA (and hence the RNA) corresponding to each gene in the cell.
In a typical DNA microarray experiment, the relative expression levels of all protein-encoding genes are compared between two states of a cell. In this type of experiment, cDNA, derived from RNA taken from the two states (typically a control and an experimental condition), is labeled with two different fluorescent molecules (Cy3 and Cy5). The labeled cDNAs are combined and hybridized to the DNA printed on the microarray. Then the microarray is washed, dried, and read using green and red lasers in a commercial scanner to excite the two fluorophores.
Using specialized computer software, the intensity of the emission peaks of both Cy3 and Cy5 fluorescent-labeled targets in each gene spot are visualized as green and red. The local area around the spot is used to determine the background value, which is typically subtracted for each target. The resulting data are then analyzed with a variety of techniques, including VxInsight, data-mining software developed at Sandia.
Werner-Washburne notes that her research “now has over $1.5 million in new funding from the National Science Foundation and National Institutes of Health that wouldn’t have been possible without the arrayer that George and our students built.”
University of New Mexico Contact: Steve Carr, scarr@unm.edu, (505) 277-1821