ALBUQUERQUE, N.M. — Sandia National Laboratories researchers witnessed molecular movements recently that could evolve into some of the first useful tools at future nanoconstruction sites, where proteins might be shuttled from place to place in tiny chemical wheelbarrows or built upon molecular scaffolding
The insights might also help create cell-sized ambulances that could travel to and selectively repair or destroy diseased cells in a human patient’s body.
Using improved observational methods, the Sandia team watched as huddled receptor — or grabber — molecules on a man-made cell membrane rapidly dispersed across the membrane when they latched onto free-floating ligands (chemical particles), then rehuddled when the ligands were removed.
The behavior mimics biological reactions at the cell level, such as immune system response to viral particles, says Darryl Sasaki of Sandia. The work is based on previous research at Sandia to create metal-detecting sensors based on chemical recognition events (https://newsreleases.sandia.gov/metal-detecting-molecules-may-find-use-in-process-water-recycling-groundwater-cleanup-virus-detection-and-more/).
“When they bind to the ligand, they each race away from their nearest neighbor,” says Sasaki. “When the ligands are removed, they race back to where they were.”
The team’s observations are published as the cover story in today’s issue of the chemical and biophysics journal Langmuir.
Portions of the work were funded by the U.S. Department of Energy’s Office of Basic Energy Sciences and through discretionary Sandia research funds.
Molecules with memories
The researchers created an artificial cell membrane made of “phospholipid bilayers” — rows of long molecules that, like empty soda bottles bobbing on water, self-organize into an orderly heads-up/tails-down formation.
They implanted this lipid film with lipids carrying tall receptor headgroups — pincher- or lasso-shaped structures that chemically grab onto free-floating ligands.
Then they watched as the receptors reacted to the addition of metal ions.
At rest the receptor-lipids pooled into aggregate zones between islands of shorter receptor-less lipids. But when metal ions (lead or copper) were added to the solution, the headgroups latched onto the ions, and ZIP!, the receptor-lipids dispersed across the membrane surface as their newly acquired electrostatic charges caused them to become mutually repulsed.
When the metal ions were removed, the wayward receptor-lipids retraced their steps and regrouped into the same aggregated pools.
The process was performed repeatedly on the same membranes with the same result — reversible reorganization.
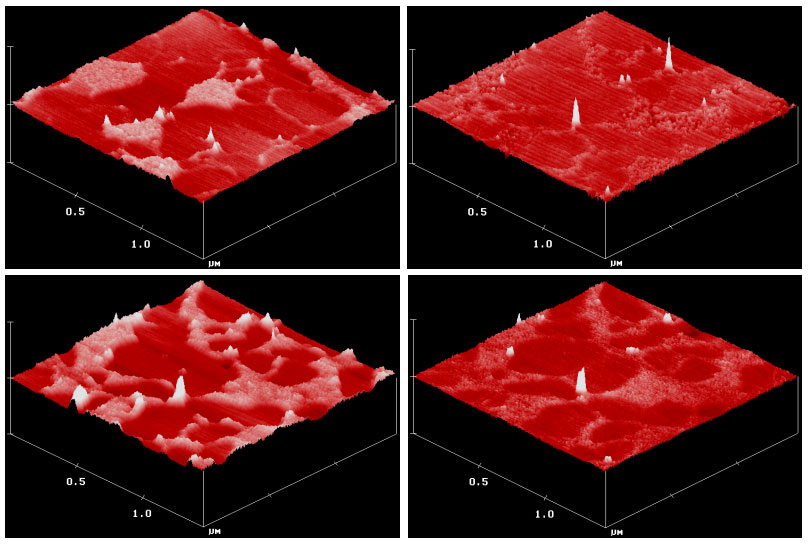
Sasaki believes the trails the receptor-lipids follow and the pools they return to correspond, quite literally, to the paths of least resistance on the membrane’s surface — areas where the lipid film is more liquid than solid, allowing the traveling lipids to flow like water.
Tracking tiny travelers
Although producing such chemical recognition events on an artificial membrane is not an achievement in itself, examining them with such fidelity is, says Sasaki. The Sandia team used novel microscopy and spectroscopic techniques to make the first documented observations of receptor-lipids dispersing and regrouping.
Fluorescent pyrene tags were attached to the tails of the receptor-lipids to aid in tracking their travels on the membrane. When the receptors were aggregated — as seen using fluorescence spectroscopy — the huddles appeared bright. When the receptors were dispersed, their fluorescent signals were dim.
In addition, the team used an atomic force microscope to map the topography of the lipid membrane, identifying locations of the tall receptor headgroups that towered 8 angstroms (about one billionth of a meter) higher than the tops of the membrane lipids.
These observations provided unprecedented clarity about the locations of the receptors in both the dispersed and aggregated states, Sasaki says.
“We’ve been able to characterize films as they change their properties at both the macroscale and nanoscale,” he says. “It’s the first time such a dynamic molecular system has been imaged this way.”
The observations will provide scientists with a better understanding of chemical recognition on cell-like membrane systems.
Nanotech’s toolbox
Perhaps more tantalizing, he says, are the possibilities the new understandings might bring to the nanotechnology community’s growing toolbox.
“The idea of using chemical recognition to form specific structures in the membrane may be a potent tool to aid in the development of controllable nanoscale architectures,” says Sasaki.
If receptor headgroups propelled to and fro by chemical recognition events can be enlisted to hoist molecules and proteins and deposit them in planned locations, he says, designing and building nanosized structures, such as single-molecule-wide wires, might be possible.
And the receptor-lipids’ tendencies to follow preferred pathways offers promise for engineered construction of nano-railroad tracks along which a variety of molecular cargos could be recurringly moved, perhaps aboard motor-protein railcars, he says.
If researchers can learn to control these routes, two- or three-dimensional lipid scaffolds might be designed upon which proteins could be laid down to build nanoscale electronic or photonic circuits. Nano-switching structures might be designed that self-construct and self-destruct based on chemical recognition events.
And researchers have long sought to build cell-like pods that, when injected into a person’s blood stream, would recognize diseased cells and release a drug to destroy those cells selectively.
“By harnessing even a fraction of the capability of cellular membrane recognition systems, it may be possible to build unique sensor systems that are not only rapid and specific in response but also are innately biocompatible,” adds Sasaki.
