ALBUQUERQUE, N.M. — COVID-19 researchers the world over face a daunting task of sifting through tens of thousands of existing coronavirus studies, looking for commonalities or data that might help in their urgent biomedical investigations.
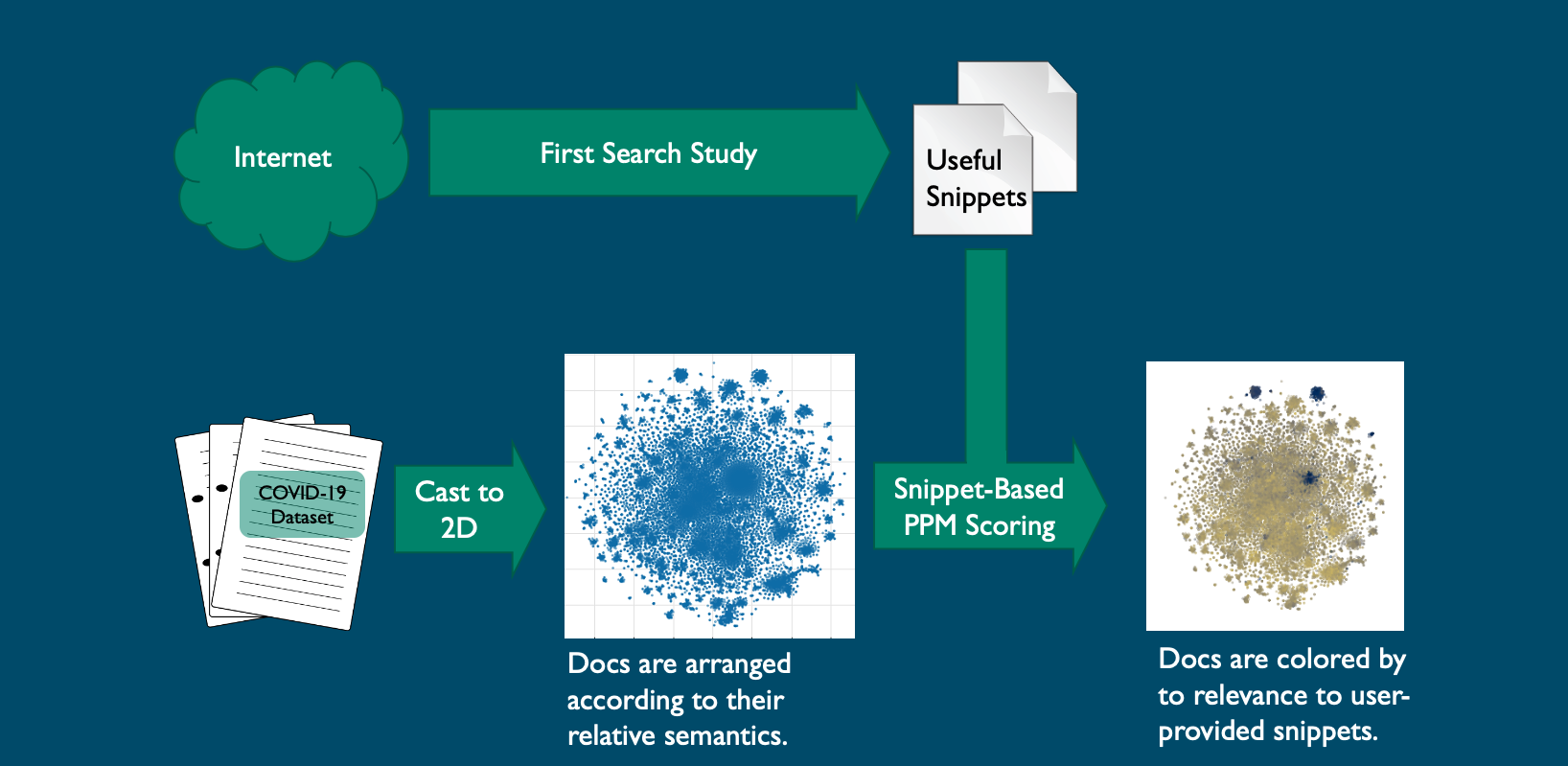
To accelerate the filtering of relevant information, Sandia National Laboratories has assembled a combination of data mining, machine-learning algorithms and compression-based analytics to bring the most useful data to the fore on an office computer. In its initial effort, investigators were able to whittle down 29,000-plus published coronavirus studies to 87 papers by identifying language and character similarities in a matter of 10 minutes. That’s rapid-response data science.
“Medical and epidemiological experts can have near-immediate access to existing pertinent research without being data scientists,” Sandia computer scientist Travis Bauer said. “With some refinement, this new process can clarify questions our public health experts need answered to fast-track COVID-19 research, particularly as new studies quickly emerge.”
The nature of rapid-response science is to quickly generate reliable results. In a seven-day effort, Sandia scientists, conceived, configured, analyzed, tested and re-analyzed an experiment helping biosecurity and public health experts isolate key coronavirus documents to rapidly access the most relevant information in defeating the COVID-19 virus.
Bauer and a team of data scientists, engineers, a human-factors expert and experts in virology, genetics, public health, biosecurity and biodefense developed and ran two different search studies — one with two experts and one with three. The experts studied “Stability of SARS-CoV-2 in aerosol droplets and other matrices,” drawn from the March 18 U.S. Department of Homeland Security master question list, intended to quickly present the current state of available information to government decisionmakers and encourage scientific discussions across the federal government.
Applying algorithms and compression data techniques Data used in the project were provided as part of a federal call to action to the tech community on a “New Machine-Readable COVID-19” dataset that, at the time, contained 29,315 research documents full of topics relevant to coronavirus. In a bid to accelerate experts’ ability to study a specific question, Sandia’s research — funded initially through the labs’ royalty income and then through the Sandia’s Laboratory Directed Research & Development program — was conducted in several stages.
In the initial stage, the study’s virology, genetics, public health, biosecurity and biodefense experts indexed the research papers and plotted that information in a two-dimensional graph using natural language processing techniques based on document content. The documents were converted into a searchable natural-language matrix and indexed or scored for searchability and relevance.
Three commonly used visualization algorithms were tested on the 29,000-document set to see which would best arrange the documents into useful clusters, Bauer said.
- The Singular Value Decomposition algorithm uncovers latent information in relationships among document terms. Bauer said, for the purposes of this study, this algorithm didn’t provide enough differentiation for a user to explore, so it was not chosen.
- The Uniform Manifold Approximation and Projection algorithm is a popular method used to broadly arrange data in two dimensions for visualization. However, for this study, UMAP as tested didn’t provide enough differentiation in the documents for experts to be able to take a deep dive into a specific COVID-19 topic. The team believed that additional tuning of this algorithm could make it more useful for this dataset.
- The T-distributed Stochastic Neighbor Embedding algorithm is a machine-learning tool that can batch similar or relevant data. The algorithm produced clearly defined collections of related information that enabled experts to explore specific COVID-19 topics. Bauer’s team determined that this algorithm could be finetuned to produce even better, more usable results.
Also in the initial phase, the same experts were asked to search for articles relevant to “Stability of SARS-CoV-2 in aerosol droplets and other matrices” using the search system or engine of their choice.
The study experts captured what they considered relevant or interesting information helpful in answering their COVID-19 question and pasted it into a Microsoft Word document. The document containing the information became the snippets that were used to create scores for articles based on how well they answered the experts’ questions.
The snippets identified included COVID-19 and coronavirus stability, case studies, test matrices and other topics. Results were plotted as points on a two-dimensional graph indicating clusters of relevant and irrelevant articles.
An analysis algorithm in the Prediction by Partial Matching data-compression technique then scored all COVID-19 documents per the snippets. Scores were used to color the documents on the two-dimensional graph, providing clusters of color that show the expert where the relevant information can be found. About 87 clustered documents were deemed highly relevant on the graph; more than 23,000 of the documents were deemed irrelevant.
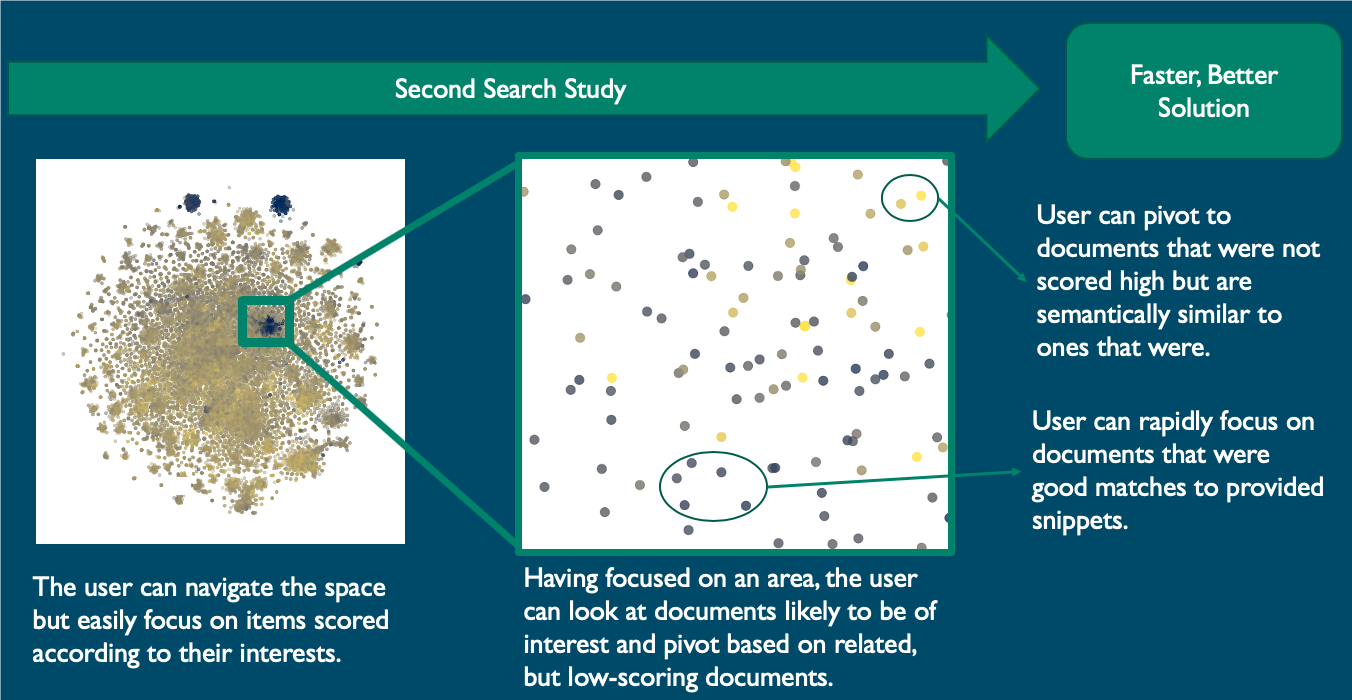
Experts in study say tools effectively categorized results, have potential Following a 30-minute session, the experts were asked to explain their search terms, how they decided which articles to view and what content they were looking for in each article.
The experts interactively explored the contrasting colored clusters that stood out as batched COVID-19-related documents. They could study any of documents to determine whether they were batched appropriately according to relevance or pivot to new snippets.
The same experts who examined the results said that the documents were accurately batched according to relevance and offered suggestions on further refining the interface by displaying information about title, authors, year, journal and abstract. The experts said they saw a lot of potential with this tool.
“Even on my office laptop computer we can sort millions of documents and make them available to the user,” Bauer said. He acknowledged that some algorithms used provided more differentiation and visual clustering, but that tuning the algorithms will improve performance.
“Technologically, it’s possible to rapidly research and adapt to experts’ needs as they work through a data set,” Bauer said. “The agility and speed with which the user interface can be developed with the right team on desktop computer systems can provide an ability to respond to specific queries quickly and adapt with the changing needs of the user.”
